Section: New Software and Platforms
OpenAlea
OpenAlea 2.0
Participants : Julien Coste, Guillaume Baty, Christophe Pradal, Christophe Godin, Frédéric Boudon, Christian Fournier.
Plant models are usually developed at different scales using various modeling paradigms: (i) imperative using a script or a compiled language, (ii) declarative to define a set of rewriting rules like in L-systems, (iii) interactive using a sketch-based interface for creating 3D models of plants, or (iv) visual programming to combine existing components.
However, all these computational paradigms have been developed in different software platforms in the plant modeling community, and, as of today, none of them provides all the modeling paradigms in an integrated software environment. However, the need to develop more complex and integrated models, often assembling many sub-models, led us to consider a modeling framework capable of supporting multiple design paradigms and models, and make them interoperable.
To address this problem we developed the OpenAlea platform. The Version 1.0 of the platform consisted of a middleware implementing a modular and component-based software architecture for assembling models written in different computer languages. OpenAlea 2.0 adds to OpenAlea 1.0 a high-level formalism dedicated to the modeling of morphogenesis that makes it possible to use several modeling paradigms (Blackboard, L-systems, Agents, Branching processes, Cellular Automata) expressed with different languages (Python, L-Py, R, Visual Porgramming, ...) to analyse and simulate shapes and their development.
It offers an integrated modeling software environment OpenAleaLab that provides users with flexible and interactive tools to combine different modeling paradigms to support the computational investigation.
OpenAleaLab
Participants : Julien Coste, Guillaume Baty, Christophe Pradal, Christophe Godin, Frédéric Boudon, Christian Fournier.
This research theme is supported by the Inria ADT OpenAlea.
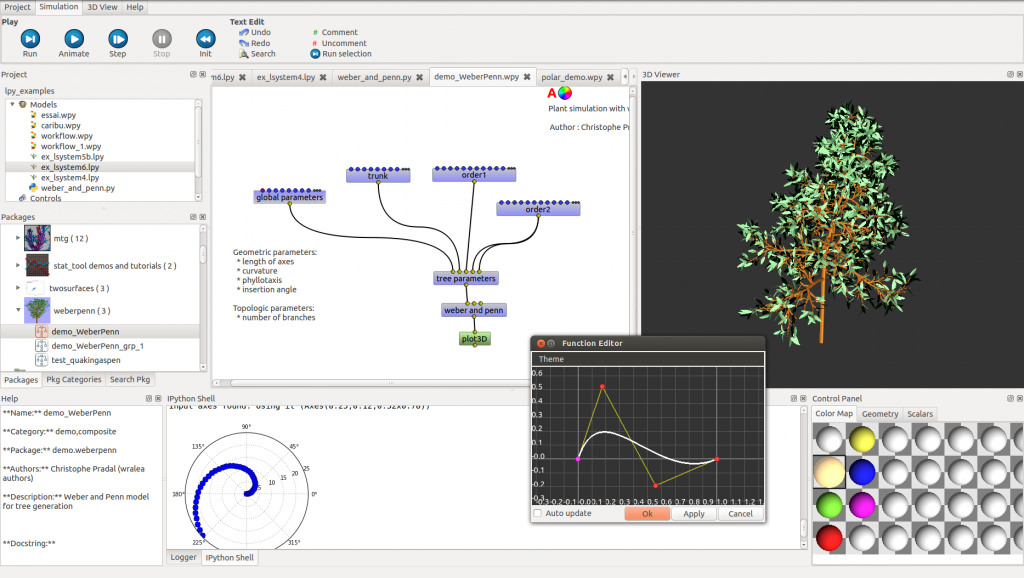
OpenAleaLab is a new integrated modeling environment (IME) for OpenAlea. This IME provides an IPython shell, a text editor, a project manager, a toolbox installer, a world data structure containing the objects and state variables shared by the different models and a 3D viewer window that makes it possible to observe the objects of the world. Different modelling paradigms, languages and tools for plant modelling are available as plug-ins, such as a visual programming environment, a L-system language, or a R editor and interpreter. OpenAleaLab is based on IPython architecture and is built using PyQt.
The core of the system is made up of a central data structure (the blackboard) called the world. This data structure may contain various computational objects that altogether define the state of the modeling system, and can be accessed (in read and write) by all the models. The investigation process can be seen as executing the system’s models in turn to explore or change dynamically the world objects.
Models are knowledge sources that can modify the world when executed. A model can call for the execution of another model as a function. In this case the model passes an input value to the called model, that inturn returns an output value. In addition it may be possible that the called model changed the world as a side effect. The user launches the execution of a first model (then referred to as the master model), which then entails recursively the hierarchical execution of all the other models downstream of it. One can see that in this framework, the execution controller is then itself considered as a model (the master model).
Similarity and Provenance in OpenAlea workflows
Participants : Sarah Cohen-Boulakia, Christophe Pradal, Moussa Yattara [IBC] , Patrick Valduriez [Inria] .
This research theme is supported by IBC and Inria.
The number of available scientific workflows, designed in OpenAlea or in other worflow systems such as Galaxy or Taverna, is increasing over time. Methods to compare the scientific workflows become a necessity, to allow duplicate detection or similarity search. Scientific workflows are complex objects, and their comparison entails a number of distinct steps from comparing atomic elements to comparison of the workflows as a whole. Various studies have implemented methods for scientific workflow comparison and came up with often contradicting conclusions upon which algorithms work best. Comparing these results is cumbersome, as the original studies mixed different approaches for different steps and used different evaluation data and metrics.
We first contribute to the field [27] by (i) comparing in isolation different approaches taken at each step of scientific workflow comparison, reporting on an number of unexpected findings, (ii) investigating how these can best be combined into aggregated measures, and (iii) making available a gold standard of over 2000 similarity ratings contributed by 15 workflow experts on a corpus of 1500 workflows and re-implementations of all methods we evaluated.
Then, we introduced a novel and intuitive workflow similarity measure that is based on layer decomposition [39] . Layer decomposition accounts for the directed dataflow underlying scientific workflows, a property which has not been adequately considered in previous methods. We comparatively evaluate our algorithm using our gold standard and show that it a) delivers the best results for similarity search, b) has a much lower runtime than other, often highly complex competitors in structure-aware workflow comparison, and c) can be stacked easily with even faster, structure-agnostic approaches to further reduce runtime while retaining result quality.
Ongoing work includes considering provenance traces of executions in the similarity metrics and augmenting the number of workflows to be shared between scientists by working on the provenance-equivalence aspects between workflows and (Python) scripts. This work will be done in the context of the IBC Young researcher grant we obtained (co-leaded by S. Cohen-Boulakia and Ch. Pradal) in collaboration with members of Zenith and the INRA phenome platform.